11 Exploratory data analysis
For this chapter you’ll need the following files, which are available for download here: ucr2017.rda and offenses_known_yearly_1960_2020.rds.
When you first start working on new data it is important to spend some time getting familiar with the data. This includes understanding how many rows and columns it has, what each row means (is each row an offender? a victim? crime in a city over a day/month/year?, etc.), and what columns it has. Basically you want to know if the data is capable of answering the question you are asking.
While not a comprehensive list, the following is a good start for exploratory data analysis of new data sets.
- What are the units (what does each row represent?)?
- What variables are available?
- What time period does it cover?
- Are there outliers? How many?
- Are there missing values? How many?
For the first part of this lesson we will use a data set of FBI Uniform Crime Reporting (UCR) data for 2017. This data includes every agency that reported their data for all 12 months of the year. In this part of the chapter we will look at some summary statistics for the variables we are interested in and make some basic graphs to visualize the data.
First, we need to load the data. Make sure your working directory is set to the folder where the data is.
load("data/ucr2017.rda")The function head() will print out the first 6 rows of every column in the data. Since we only have 9 columns, we will use this function. Be careful when you have many columns (100+) as printing all of them out makes it difficult to read.
head(ucr2017)
# ori year agency_name state population actual_murder actual_rape_total
# 1 AK00101 2017 anchorage alaska 296188 27 391
# 2 AK00102 2017 fairbanks alaska 32937 10 24
# 3 AK00103 2017 juneau alaska 32344 1 50
# 4 AK00104 2017 ketchikan alaska 8230 1 19
# 5 AK00105 2017 kodiak alaska 6198 0 15
# 6 AK00106 2017 nome alaska 3829 0 7
# actual_robbery_total actual_assault_aggravated
# 1 778 2368
# 2 40 131
# 3 46 206
# 4 0 14
# 5 4 41
# 6 0 52From these results it appears that each row is a single agency’s annual data for 2017, and the columns show the number of crimes for four crime categories included.
Finally, we can run names() to print out every column name. We can already see every name from head(), but this is useful when we have many columns and don’t want to use head().
names(ucr2017)
# [1] "ori" "year"
# [3] "agency_name" "state"
# [5] "population" "actual_murder"
# [7] "actual_rape_total" "actual_robbery_total"
# [9] "actual_assault_aggravated"11.1 Summary and Table
An important function in understanding the data you have is summary() which, as discussed in Section 2.5, provides summary statistics on the numeric columns you have. Let’s take a look at the results before seeing how to do something similar for categorical columns.
summary(ucr2017)
# ori year agency_name state
# Length:15764 Min. :2017 Length:15764 Length:15764
# Class :character 1st Qu.:2017 Class :character Class :character
# Mode :character Median :2017 Mode :character Mode :character
# Mean :2017
# 3rd Qu.:2017
# Max. :2017
# population actual_murder actual_rape_total actual_robbery_total
# Min. : 0 Min. : 0.000 Min. : -2.000 Min. : -1.00
# 1st Qu.: 914 1st Qu.: 0.000 1st Qu.: 0.000 1st Qu.: 0.00
# Median : 4460 Median : 0.000 Median : 1.000 Median : 0.00
# Mean : 19872 Mean : 1.069 Mean : 8.262 Mean : 19.85
# 3rd Qu.: 15390 3rd Qu.: 0.000 3rd Qu.: 5.000 3rd Qu.: 4.00
# Max. :8616333 Max. :653.000 Max. :2455.000 Max. :13995.00
# actual_assault_aggravated
# Min. : -1.00
# 1st Qu.: 1.00
# Median : 5.00
# Mean : 49.98
# 3rd Qu.: 21.00
# Max. :29771.00The table() function returns every unique value in a category and how often that value appears. Unlike summary() we can’t just put the entire data set into the (), we need to specify a single column. To specify a column you use the dollar sign notation, which is data$column. For most functions we use to examine the data as a whole, such as head(), you can do the same for a specific column.
head(ucr2017$agency_name)
# [1] "anchorage" "fairbanks" "juneau" "ketchikan" "kodiak" "nome"There are only two columns in our data with categorical values that we can use - year and state, so let’s use table() on both of them. The columns ori and agency_name are also categorical but as each row of data has a unique ORI and name, running table() on those columns would not be helpful.
table(ucr2017$year)
#
# 2017
# 15764We can see that every year in our data is 2017, as expected based on the data name. year is a numerical column so why can we use table() on it? R doesn’t differentiate between numbers and characters when seeing how often each value appears. If we ran table() on the column “actual_murder” it would tell us how many times each unique value in the column appeared in the data. That wouldn’t be very useful as we don’t really care how many times an agency has, for example, 7 murders. As numeric variables often have many more unique values than character variables, it also leads to many values being printed, making it harder to understand. For columns where the number of categories is important to us, such as years, states, neighborhoods, we should use table().
table(ucr2017$state)
#
# alabama alaska arizona
# 305 32 107
# arkansas california colorado
# 273 732 213
# connecticut delaware district of columbia
# 107 63 3
# florida georgia guam
# 603 522 1
# hawaii idaho illinois
# 4 95 696
# indiana iowa kansas
# 247 216 309
# kentucky louisiana maine
# 352 192 135
# maryland massachusetts michigan
# 152 346 625
# minnesota mississippi missouri
# 397 71 580
# montana nebraska nevada
# 108 225 59
# new hampshire new jersey new mexico
# 176 576 116
# new york north carolina north dakota
# 532 310 108
# ohio oklahoma oregon
# 532 409 172
# pennsylvania rhode island south carolina
# 1473 49 427
# south dakota tennessee texas
# 92 466 999
# utah vermont virginia
# 125 85 407
# washington west virginia wisconsin
# 250 200 433
# wyoming
# 57This shows us how many times each state is present in the data. States with a larger population tend to appear more often; this makes sense as those states have more agencies to report. Right now the results are in alphabetical order, but when knowing how frequently something appears, we usually want it ordered by frequency. We can use the sort() function to order the results from table(). Just put the entire table() function inside of the () in sort().
sort(table(ucr2017$state))
#
# guam district of columbia hawaii
# 1 3 4
# alaska rhode island wyoming
# 32 49 57
# nevada delaware mississippi
# 59 63 71
# vermont south dakota idaho
# 85 92 95
# arizona connecticut montana
# 107 107 108
# north dakota new mexico utah
# 108 116 125
# maine maryland oregon
# 135 152 172
# new hampshire louisiana west virginia
# 176 192 200
# colorado iowa nebraska
# 213 216 225
# indiana washington arkansas
# 247 250 273
# alabama kansas north carolina
# 305 309 310
# massachusetts kentucky minnesota
# 346 352 397
# virginia oklahoma south carolina
# 407 409 427
# wisconsin tennessee georgia
# 433 466 522
# new york ohio new jersey
# 532 532 576
# missouri florida michigan
# 580 603 625
# illinois california texas
# 696 732 999
# pennsylvania
# 1473And if we want to sort it in decreasing order of frequency, we can use the parameter decreasing in sort() and set it to TRUE. A parameter is just an option used in an R function to change the way the function is used or what output it gives. Almost all functions have these parameters, and they are useful if you don’t want to use the default setting in the function. This parameter, decreasing, changes the sort() output to print from largest to smallest. By default this parameter is set to FALSE, and here we say it is equal to TRUE.
sort(table(ucr2017$state), decreasing = TRUE)
#
# pennsylvania texas california
# 1473 999 732
# illinois michigan florida
# 696 625 603
# missouri new jersey new york
# 580 576 532
# ohio georgia tennessee
# 532 522 466
# wisconsin south carolina oklahoma
# 433 427 409
# virginia minnesota kentucky
# 407 397 352
# massachusetts north carolina kansas
# 346 310 309
# alabama arkansas washington
# 305 273 250
# indiana nebraska iowa
# 247 225 216
# colorado west virginia louisiana
# 213 200 192
# new hampshire oregon maryland
# 176 172 152
# maine utah new mexico
# 135 125 116
# montana north dakota arizona
# 108 108 107
# connecticut idaho south dakota
# 107 95 92
# vermont mississippi delaware
# 85 71 63
# nevada wyoming rhode island
# 59 57 49
# alaska hawaii district of columbia
# 32 4 3
# guam
# 111.2 Graphing
We often want to make quick plots of our data to get a visual understanding of the data. We will learn a different - and in my opinion a superior - way to make graphs in Chapters 14 and 15, but for now let’s use the function plot(). The plot() function is built into R so we don’t need to use any packages for it.
Let’s make a few scatterplots showing the relationship between two variables. With plot() the syntax (how you write the code) is plot(x_axis_variable, y_axis_variable). So all we need to do is give it the variable for the x- and y-axis. Each dot will represent a single agency (a single row in our data).
plot(ucr2017$actual_murder,
ucr2017$actual_robbery_total)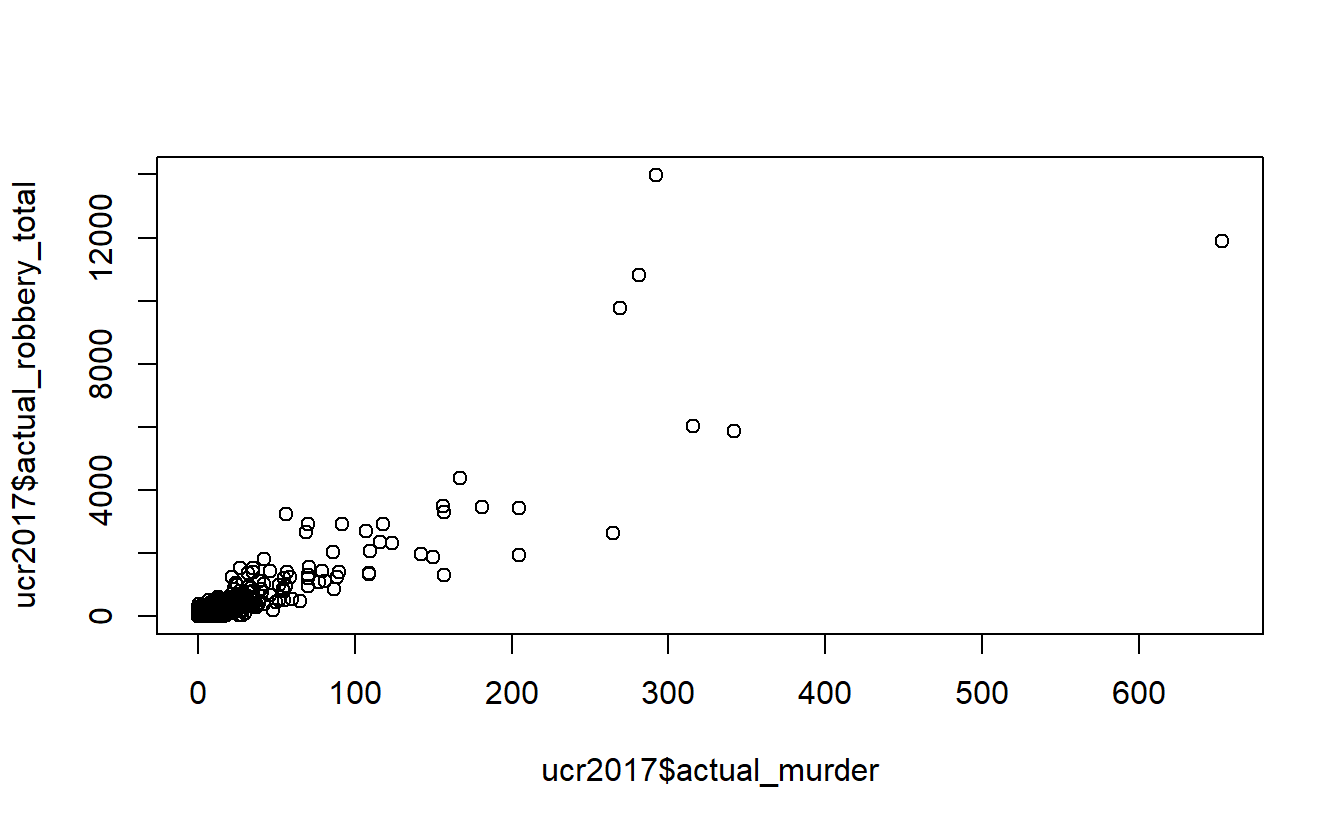
Above we are telling R to plot the number of murders on the x-axis and the number of robberies on the y-axis. This shows the relationship between a city’s number of murders and number of robberies. We can see that there is a relationship where more murders is correlated with more robberies. However, there are a huge number of agencies in the bottom-left corner that have very few murders or robberies. This makes sense as - as we see in the summary() above - most agencies are small, with the median population under 5,000 people.
To try to avoid that clump of small agencies at the bottom, let’s make a new data set of only agencies with a population over 1 million. We will use the filter() function from the dplyr package that was introduced in Chapter 10. For filter(), we need to first include our data set name, which is ucr2017, and then say our conditional statement. Our conditional statement is that rows in the “population” column have a value of over 1 million. For the dplyr functions we don’t put our column name in quotes.
And we’ll assign our results to a new object called ucr2017_big_cities Since we’re using the dplyr package we need to tell R that we want to use it by using library(dplyr).
library(dplyr)
#
# Attaching package: 'dplyr'
# The following objects are masked from 'package:stats':
#
# filter, lag
# The following objects are masked from 'package:base':
#
# intersect, setdiff, setequal, union
ucr2017_big_cities <- filter(ucr2017, population > 1000000) Now we have 18 agencies with a population of over 1 million people.
Now we can do the same graph as above but using this new data set.
plot(ucr2017_big_cities$actual_murder,
ucr2017_big_cities$actual_robbery_total)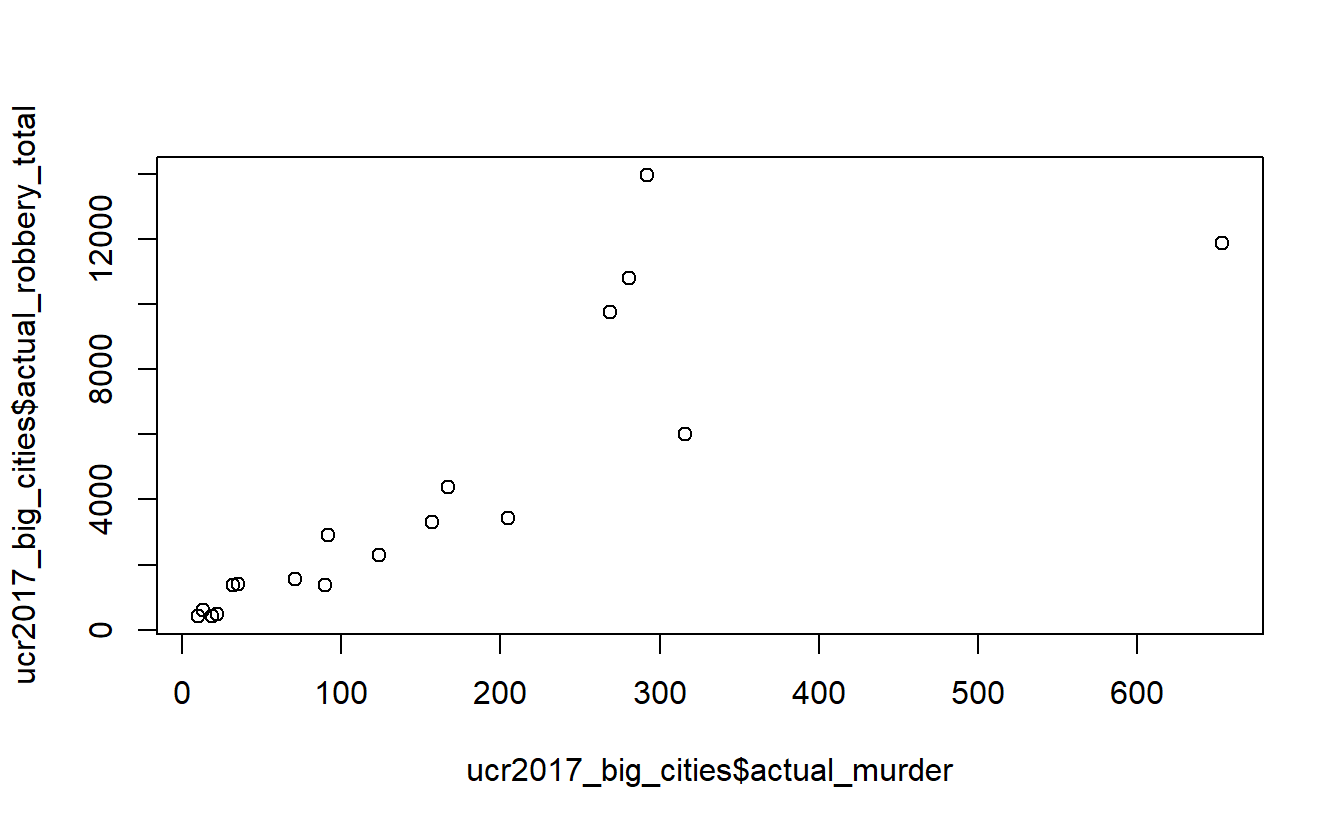
The problem is somewhat solved. There is still a small clumping of agencies with few robberies or murders, but the issue is much better. And interestingly the trend is similar with this small subset of data as with all agencies included.
To make our graph look better, we can add labels for the axes and a title (there are many options for changing the appearance of this graph, we will just use these three).
- xlab - X-axis label
- ylab - Y-axis label
- main - Graph title
Like all parameters, we add them in the () of plot() and separate each parameter by a comma. Since we are adding text to write in the plot, all of these parameter inputs must be in quotes.
plot(ucr2017_big_cities$actual_murder,
ucr2017_big_cities$actual_robbery_total,
xlab = "Murders",
ylab = "Robberies",
main = "Relationship between murder and robbery")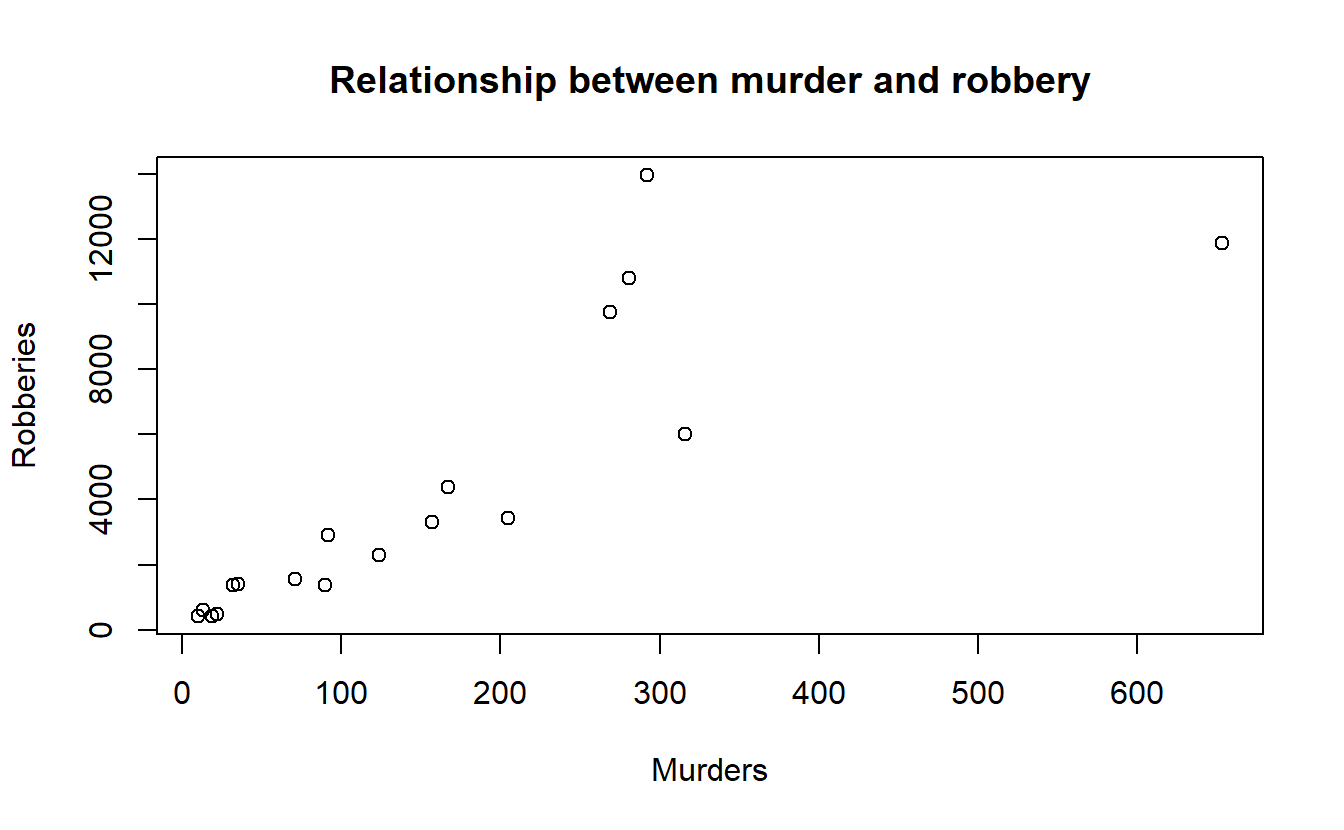
11.3 Aggregating (summaries of groups)
Right now we have the number of crimes in each agency. For many policy analyses we’d be looking at the effect on the state as a whole, rather than at the agency-level. If we wanted to do this in our data, we would need to aggregate up to the state level. Aggregating data means that we group values at some higher level than they currently are (e.g. from agency to state, from day to month, from city street to city neighborhood) and then do some mathematical operation of our choosing (in our case usually sum) to that group.
In Section 10.3.3 we started to see if marijuana legalization affected murder in Colorado. We subsetted the data to only include agencies in Colorado from 2011-2017. Now we can continue to answer the question by aggregating to the state-level to see the total number of murders per year.
Let’s think about how our data are and how we would (theoretically, before we write any code) find that out.
Our data has a single row for each agency, and we have a column indicating the year the agency reported. So how would we find out how many murders happened in Colorado for each year? Well, first we take all the agencies in 2011 (the first year we’re looking at) and add up the murders for all agencies that reported that year. Then take all the rows in 2012 and add up their murders. And so on for all the years.
To do this in R, we’ll be using two new functions from the dplyr package: group_by() and summarize().
These functions do the aggregation process in two steps. First we use group_by() to tell R which columns we want to group our data by - these are the higher level of aggregation columns so in our case will be the year of data. Then we need to sum up the number of murders each year. We do this using summarize(), and we’ll specify in the function that we want to sum up the data, rather than use some other math operation on it like finding the average number of murders each year.
First, let’s load back in the data and then repeat the subsetting code we did in Chapter 10.3.3 to keep only data for Colorado from 2011 through 2017. We’ll also include the “actual_robbery_total” column that we excluded in Chapter 10.3.3 so we can see how easy it is to aggregate multiple columns at once using this method.
ucr <- readRDS("data/offenses_known_yearly_1960_2020.rds")
colorado <- filter(ucr, state == "colorado",
year %in% 2011:2017)
colorado <- select(colorado, actual_murder, actual_robbery_total,
state, year, population, ori, agency_name)First we must group the data by using the group_by() function. Here we’re just grouping the data by year, but we could group it by multiple columns if we want by adding a comma and then the next column we want. Following other dplyr function syntax, we first input the data set name and then the column name - neither of which need to be in quotes.
colorado <- group_by(colorado, year)Now we can summarize the data using the summarize() function. As with other dplyr functions the first input is the data set name. Then we choose our math function (sum, mean, median, etc.) and just apply that function on the column we want. So in our case we want the sum of murders so we use sum() and include the column we want to aggregate inside of sum()’s parentheses.
summarize(colorado, sum(actual_murder))
# # A tibble: 7 × 2
# year `sum(actual_murder)`
# <dbl> <dbl>
# 1 2011 154
# 2 2012 163
# 3 2013 172
# 4 2014 148
# 5 2015 173
# 6 2016 203
# 7 2017 218If we want to aggregate another column we just add a comma after our initial column and add another math operation function and the column we want. Here we’re also using sum(), but we could use different math operations if we want - they don’t need to be the same.
summarize(colorado, sum(actual_murder),
sum(actual_robbery_total))
# # A tibble: 7 × 3
# year `sum(actual_murder)` `sum(actual_robbery_total)`
# <dbl> <dbl> <dbl>
# 1 2011 154 3287
# 2 2012 163 3369
# 3 2013 172 3122
# 4 2014 148 3021
# 5 2015 173 3305
# 6 2016 203 3513
# 7 2017 218 3811We could even do different math operations on the same column and we’d get multiple columns from it. Let’s add another column showing the average number of robberies as an example.
summarize(colorado, sum(actual_murder),
sum(actual_robbery_total),
mean(actual_robbery_total))
# # A tibble: 7 × 4
# year `sum(actual_murder)` `sum(actual_robbery_total)` mean(actual_robbery_t…¹
# <dbl> <dbl> <dbl> <dbl>
# 1 2011 154 3287 11.2
# 2 2012 163 3369 11.2
# 3 2013 172 3122 10.3
# 4 2014 148 3021 9.94
# 5 2015 173 3305 10.9
# 6 2016 203 3513 11.6
# 7 2017 218 3811 12.5
# # … with abbreviated variable name ¹`mean(actual_robbery_total)`By default summarize() calls the columns it makes using what we include in the parentheses. Since we said “sum(actual_murder)”, to get the sum of the murder column, it names that new column “sum(actual_murder)”. Usually we’ll want to name the columns ourselves. We can do this by assigning the summarized column to a name using “name =” before it. For example, we could write “murders = sum(actual_murder)” and it will name that column “murders” instead of “sum(actual_murder)”. Like other things in dplyr functions, we don’t need to put quotes around our new column name. We’ll assign this final summarized data to an object called “colorado_agg” so we can use it to make graphs. And to be able to create crime rates per population, we’ll also find the sum of the population for each year.
colorado_agg <- summarize(colorado,
murders = sum(actual_murder),
robberies = sum(actual_robbery_total),
population = sum(population))
colorado_agg
# # A tibble: 7 × 4
# year murders robberies population
# <dbl> <dbl> <dbl> <dbl>
# 1 2011 154 3287 5155993
# 2 2012 163 3369 5227884
# 3 2013 172 3122 5308236
# 4 2014 148 3021 5402555
# 5 2015 173 3305 5505856
# 6 2016 203 3513 5590124
# 7 2017 218 3811 5661529Now we can see that the total number of murders increased over time. So can we conclude that marijuana legalization increases murder? No, all this analysis shows is that the years following marijuana legalization, murders increased in Colorado. But that can be due to many reasons other than marijuana. For a proper analysis you’d need a comparison state that is similar to Colorado prior to legalization (and that didn’t legalize marijuana) and see if their murders changes following Colorado’s legalization.
To control for population, we’ll standardize our murder data by creating a murder rate per 100,000 people. We can do this by dividing the murder column by the population column and then multiplying by 100,000. Let’s do that and assign the result into a new column called “murder_rate”.
colorado_agg$murder_rate <- colorado_agg$murders /
colorado_agg$population * 100000If we also wanted a robbery rate we’d do the same with the robberies column.
colorado_agg$robbery_rate <- colorado_agg$robberies /
colorado_agg$population * 100000The dplyr package has a helpful function that can do this too, and allows us to do it while writing less code. The mutate() function lets us create or alter columns in our data. Like other dplyr functions we start by including our data set in the parentheses, and then we can follow standard assignment (covered in Section 2.2) though we must use = here and not <-. A benefit of using mutate() is that we don’t have to write out our data set name each time. So we’d write murder_rate = murders / population * 100000. And if we wanted to make two (or more) columns at the same time we just add a comma after our first assignment and then do the next assignment.
mutate(colorado_agg,
murder_rate = murders / population * 100000,
robbery_rate = robberies / population * 100000)
# # A tibble: 7 × 6
# year murders robberies population murder_rate robbery_rate
# <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
# 1 2011 154 3287 5155993 2.99 63.8
# 2 2012 163 3369 5227884 3.12 64.4
# 3 2013 172 3122 5308236 3.24 58.8
# 4 2014 148 3021 5402555 2.74 55.9
# 5 2015 173 3305 5505856 3.14 60.0
# 6 2016 203 3513 5590124 3.63 62.8
# 7 2017 218 3811 5661529 3.85 67.3Now let’s make a plot of this data showing the murder rate over time. With time-series graphs we want the time variable to be on the x-axis and the numeric variable that we are measuring to be on the y-axis.
plot(x = colorado_agg$year,
y = colorado_agg$murder_rate)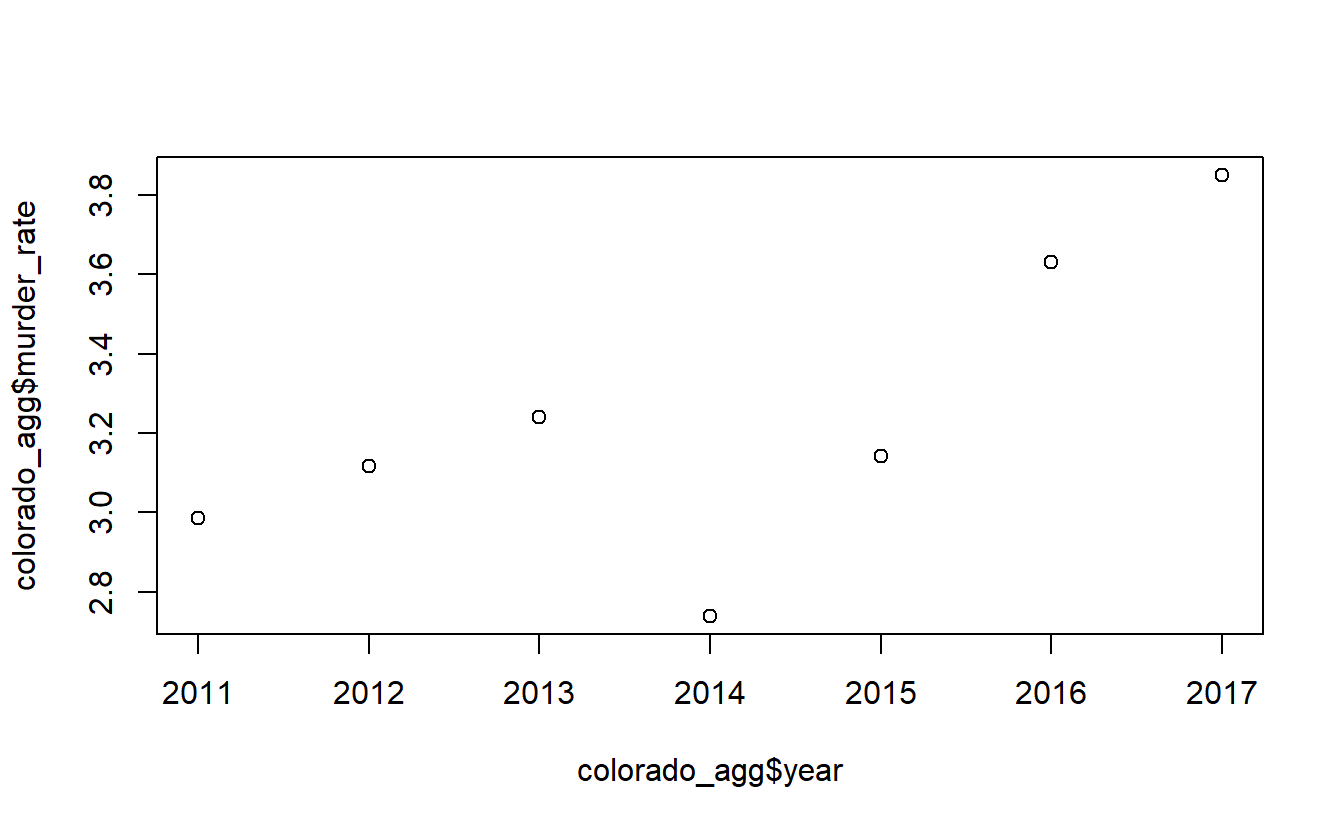
By default plot() makes a scatterplot. If we set the parameter type to “l” it will be a line plot.
plot(x = colorado_agg$year,
y = colorado_agg$murder_rate,
type = "l")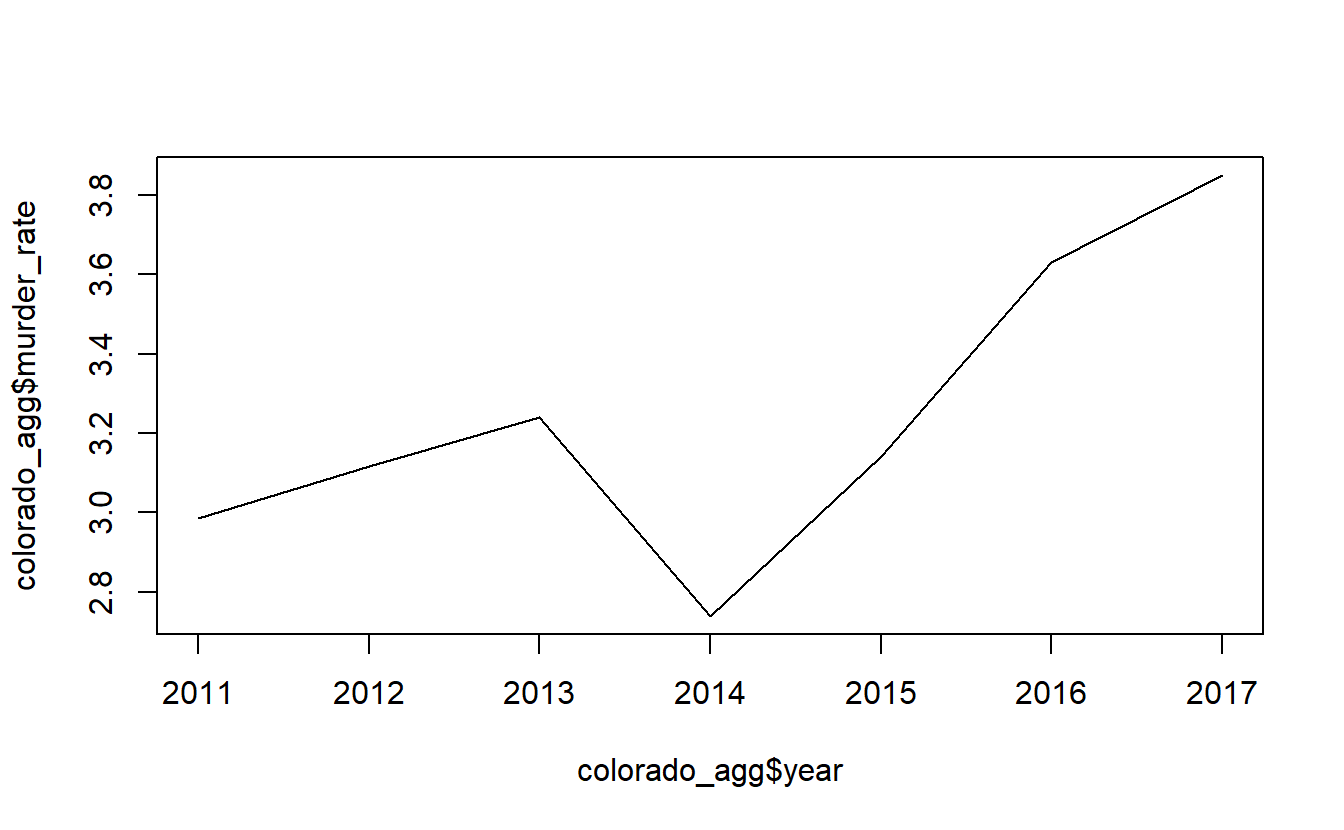
We can add some labels and a title to make this graph easier to read.
plot(x = colorado_agg$year,
y = colorado_agg$murder_rate,
type = "l",
xlab = "Year",
ylab = "Murders per 100k Population",
main = "Murder Rate in Colorado, 2011-2017")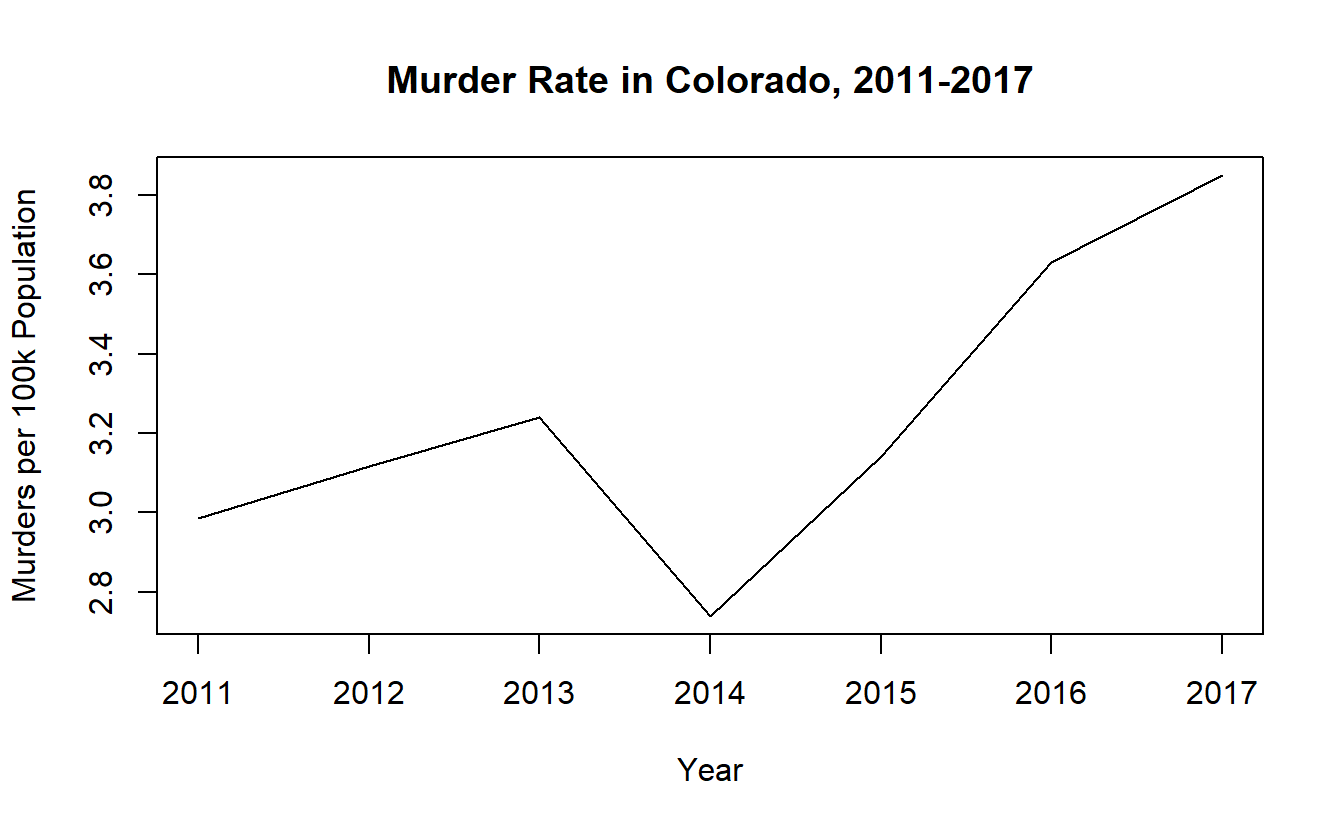
11.4 Pipes in dplyr
To end this chapter we’ll talk about something called a pipe that is a very useful and powerful part of dplyr.
Think about the math equation 1 + 2 + 3 + 4. Here we know that we add 1 and 2 together, and then add the result to 3 and then add the result of that to 4. This is much simpler to write than splitting everything up and summing each value together in a different line. In terms of R, we have so far been doing things as if we could only add two numbers together and then need a separate line to add the third (and another line to add the fourth) number. For example, below are the two lines of code we used to subset the data to just the right state and years we wanted, and the columns we wanted. We did this in two separate lines. In our math example, we did 1 + 2. And then found the answer, and separately did 3 + 3. And then again found the answer and did 6 + 4.
colorado <- filter(ucr, state == "colorado",
year %in% 2011:2017)
colorado <- select(colorado,
actual_murder,
actual_robbery_total,
state,
year,
population,
ori,
agency_name)
head(colorado)
# actual_murder actual_robbery_total state year population ori
# 1 7 80 colorado 2017 99940 CO00100
# 2 11 93 colorado 2016 100526 CO00100
# 3 6 68 colorado 2015 100266 CO00100
# 4 6 58 colorado 2014 98569 CO00100
# 5 7 44 colorado 2013 97146 CO00100
# 6 7 55 colorado 2012 93542 CO00100
# agency_name
# 1 adams
# 2 adams
# 3 adams
# 4 adams
# 5 adams
# 6 adamsWith dplyr we actually do have a way to chain together functions; to do the programming equivalent of 1 + 2 + 3 + 4 all at once.17 We do this through what is called a pipe, which allows us to take the result of one function and immediately put it into another function without having to save the initial result or start a new line of code. To use a pipe we put the following code after the end of a function: %>%.
These three characters, %>%, are the pipe, and they must be written exactly like this. The pipe is itself actually a function, but it is a special type of function we won’t go into detail about. Personally I don’t think this really looks like a pipe at all, but it is called a pipe so that’s the terminology I’ll be using. How a pipe technically works is that it takes the output of the initial function (which is usually a tibble, which is the tidyverse’s modified version of a data.frame) and puts it automatically in the first input in the next function. This won’t work for all functions but nearly all functions from the tidyverse collection of packages have a data set as the first input so it will work here. The benefit is that we don’t need to keep saving the output from functions or specifying which data set to include in each function.
As an example, we’ll rewrite the previous code using a pipe. We start with our data.frame, which is normally the first thing we put in any dplyr function, and then immediately have a pipe %>% into a dplyr function, which here is filter(). Now we don’t need to say what the data set is because it takes the last thing that was piped into the function, which in our case is the entire data.frame ucr. After our filter() is done we have another pipe and go into select(). Now select() will use as its first input whatever is outputted from the filter(). So the input to select() will be the subsetted data output from filter(). We can have as many pipes as we wish, and chain many different dplyr functions together, but we just use two functions here so we’ll end after our select() function.
colorado <- ucr %>% filter(state == "colorado",
year %in% 2011:2017) %>%
select(actual_murder,
actual_robbery_total,
state, year, population,
ori, agency_name)If we check results using head(), we can see that this code is exactly the same as not using pipes.
head(colorado)
# actual_murder actual_robbery_total state year population ori
# 1 7 80 colorado 2017 99940 CO00100
# 2 11 93 colorado 2016 100526 CO00100
# 3 6 68 colorado 2015 100266 CO00100
# 4 6 58 colorado 2014 98569 CO00100
# 5 7 44 colorado 2013 97146 CO00100
# 6 7 55 colorado 2012 93542 CO00100
# agency_name
# 1 adams
# 2 adams
# 3 adams
# 4 adams
# 5 adams
# 6 adamsThe normal way to write code using pipes is to have a new line after the pipe and after each comma in filter() and select(). This doesn’t change how the code works at all, but it is easier to read now because it has less code bunched together in a single line.
colorado <- ucr %>%
filter(state == "colorado",
year %in% 2011:2017) %>%
select(actual_murder,
actual_robbery_total,
state,
year,
population,
ori,
agency_name)Pipes are technically from the
magrittrpackage, but we’ll just be using pipes in the context of using functions fromdplyror other tidyverse packages.↩︎